Researchers analyzed data on three variants of concern and found that viral loads were higher and infection lasted longer for the B.1.1.7 variant. They all were more infectious than the original strain, and transmissibility also depended on population demographics.
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has evolved since it first emerged in late 2019. Several mutations of the virus have since been observed, with some variants appearing more infectious than the original strain, which also appear to evade immunity gained by previous infections.
Some of these variants have been labeled as variants of concern (VOC) because of their greater transmissibility and infectiousness. These have also been associated with second and third waves in the infections in many countries, leading to their close monitoring, either by sequencing the complete viral genome or reverse-transcription polymerase chain reaction (RT-PCR).
Some RT-PCR studies have suggested that the VOCs may be causing higher viral loads, but those studies were done only at one-time point. In a new study published on the medRxiv* preprint server, researchers from France report how different variants affect virus kinetics in infections.
Testing virus loads
The researchers used data from RT-PCR tests specific to variants performed between February and April 2021 on SARS-CoV-2 positive samples. They used data from more than 17,000 samples. About 73% of the samples were of the B.1.1.7 variant, 6% of the B.1.351 or P.1 variants, and the rest were wild-type.
A model was developed that included several co-factors, such as the patient's age, the virus lineage, and whether the patient was hospitalized.
They found that the viral loads were different between the wild-type virus and the different variants. For the B.1.1.7 strain, peak virus load increased with age. The cycle threshold values for the B.1.351 and P.1 strains were slightly lower than that for the wild-type strain, indicating slightly lower viral loads for these variants.
In addition, the rate of decrease of viral load with time was lower for the B.1.1.7 strain compared to the wild-type virus. The data also showed higher peak loads and a lower rate of viral load decrease in hospitalized individuals, about 1.5 days more.
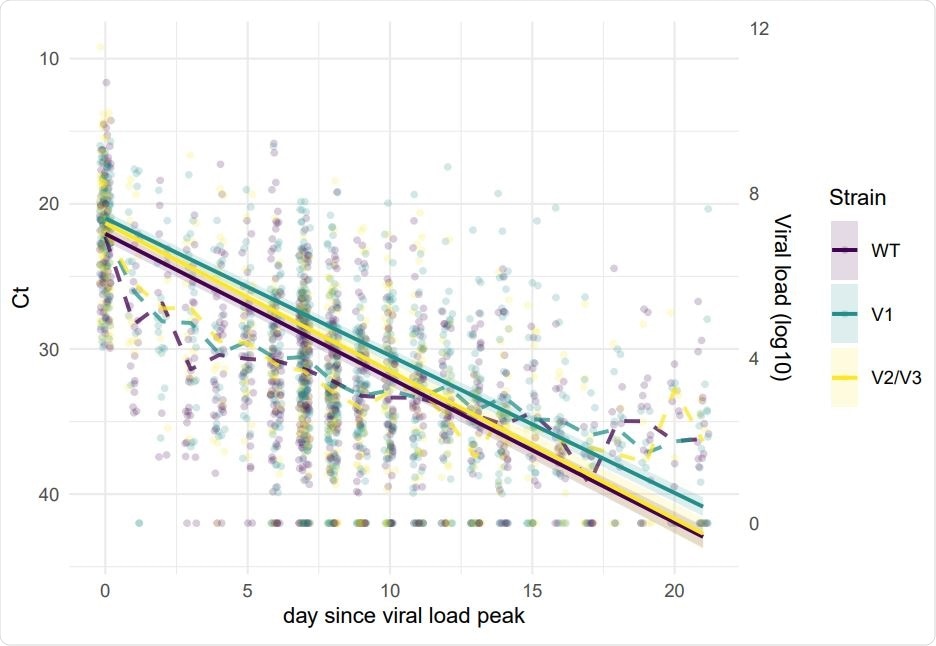
Variants more infectious
There was a significant correlation between the cycle threshold value and the decrease in viral loads, suggesting a linear relationship between the cycle threshold value and infectiousness. These researchers found there was a greater risk of transmission for variants than for wild-type viruses.
For the B.1.1.7 strain, greater transmission was more evident in countries with an older population like Japan (a median advantage of 41%) and France. In contrast, for the B.1.351 and P.1 variants were more evident for countries with a younger population like Niger (median advantage of 26%).
Thus, the analysis shows the SARS-CoV-2 variants have a higher potential of transmission, which also depends on the population's demographics. The authors suggest future studies should look at combing the cycle threshold data and the serological data to understand how the B.1.351 and P.1 variants escape our immune response.
*Important Notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Elie, E. et al. (2021) Inferring SARS-CoV-2 variant within-host kinetics. medRxiv, https://doi.org/10.1101/2021.05.26.21257835, https://www.medrxiv.org/content/10.1101/2021.05.26.21257835v1
Posted in: Medical Research News | Disease/Infection News
Tags: Coronavirus, Coronavirus Disease COVID-19, CT, Genome, Hospital, Immune Response, Polymerase, Polymerase Chain Reaction, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Transcription, Virus

Written by
Lakshmi Supriya
Lakshmi Supriya got her BSc in Industrial Chemistry from IIT Kharagpur (India) and a Ph.D. in Polymer Science and Engineering from Virginia Tech (USA).
Source: Read Full Article
