
Rice University synthetic biologists have developed a new way to study a sensory system that salmonella, E. coli and other pathogens use to sicken millions of people each year.
In pathogenic bacteria, the sensor PhoPQ turns on a cluster of genes required for infection. Rice researchers rewired PhoPQ to a reporter gene that made a fluorescent protein each time the gene was stimulated. The researchers then exposed their engineered microbes to thousands of small proteins called peptides, and the microbes emitted a telltale fluorescent glow each time a peptide triggered the PhoPQ sensor.
In a paper published in Nature Chemical Biology, Rice’s Jeff Tabor, lead co-author Kathryn Brink and colleagues describe their technique and show how it can be combined with a method called peptide display to rapidly scan large libraries of peptides and identify those that activate bacterial sensors.
“This is the first high-throughput method to study peptide-receptor interactions in bacteria,” Tabor said.
Peptides are an important class of activators of bacterial sensor receptors. Bacteria can broadcast peptides like chemical tweets to warn of danger or organize collective action. Pathogenic bacteria have also evolved to detect threatening peptides, including human antimicrobial peptides (AMPs).
“AMPs are small proteins our bodies produce to fight infections by killing bacteria,” Tabor said. “They are made by the immune system and many tissues. They can also play ‘double duty’ by serving as signals that activate or repress immune activity, like inflammation, which kills bacteria.”
Brink, Tabor, Rice graduate student Maxwell Hunt and their colleagues used plasmids to engineer E. coli to display AMPs on their outer surface.
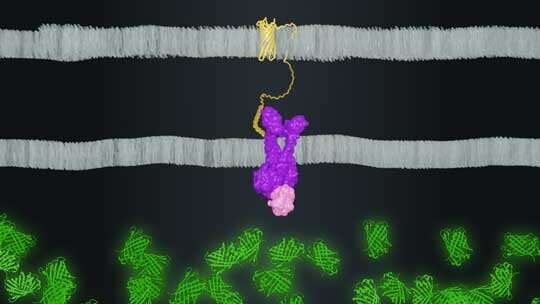
“The benefits of this method are that we can easily ‘treat’ E. coli with thousands of different peptides by building thousands of plasmids,” Tabor said. “That is cheap and easy compared to chemical peptide synthesis.”
Previous studies had shown PhoPQ sensed one human AMP as well as several human signaling peptides that play a role in inflammation.
“Large-scale chemical synthesis of peptides is prohibitively expensive, so the previous record for the number of peptides studied for the ability to activate PhoPQ was nine,” Tabor said. “By encoding peptides genetically, we were able to screen more than 3,000 in a single experiment.”
He said the experiment generated “a mountain of data, and hidden within that was information about the general properties of peptides PhoPQ senses.”
Brink and Tabor used machine learning, a form of artificial intelligence, to analyze the data, find hidden patterns and discover “new ‘rules’ for peptide features that are needed to activate PhoPQ,” he said.
“The human body produces about 140 AMPs,” Tabor said. “Of those, only two had ever been tested for their ability to activate PhoPQ. We tested most of them and discovered 13 new human AMPs that activate this bacterial sensor. Those are produced in different tissues of the body, including the small intestine, urinary tract and immune cells. That suggests these bacteria are defending themselves against many more of our immune system’s attacks, and potentially in more places in our bodies, than was previously thought.”
PhoPQ is a highly conserved genetic pathway in bacterial pathogens. To broaden their findings and explore how PhoPQ sensing evolved in other species, Brink and Tabor also characterized the response of PhoPQ from both pathogenic and nonpathogenic strains of E. coli and another pathogen called Klebsiella pneumoniae.
“We found striking evidence that PhoPQ in pathogenic E. coli and K. pneumoniae responds more strongly to AMPs that are enriched in tissues they infect,” Tabor said. For instance, the PhoPQ from pathogenic E. coli responded more strongly to AMPs in the bladder, where that organism frequently causes infections.
“This result suggests there may be an arms race where humans evolve new AMPs to attack bacteria in new ways, and bacteria evolve to specifically defend against those AMPs,” Tabor said.
He said the new peptide display method is not compatible with all peptides.
“Some have advanced chemical modifications that can’t be made using our system,” Tabor said. “But our method is compatible with many peptides, including those with disulfide bonds that are present in some AMPs. We think our method could be used to discover peptides that activate other bacterial receptors involved in virulence, interbacterial warfare, biofilm formation, infection and other processes in many species of bacteria.”
Tabor is a professor of bioengineering and of biosciences. Brink is a postdoctoral fellow at Stanford University’s School of Medicine and an alumna of Rice’s systems, synthetic and physical biology Ph.D. program.
Additional study co-authors include Maxwell Hunt, Andrew Mu, Kevin Lorch and Brett Pogostin, all of Rice; Ken Groszman of the Massachusetts Institute of Technology; and Ky Hoang and John Gunn of Ohio State University.
More information:
Kathryn R. Brink et al, An E. coli display method for characterization of peptide-sensor kinase interactions, Nature Chemical Biology (2022). DOI: 10.1038/s41589-022-01207-z
Journal information:
Nature Chemical Biology
Source: Read Full Article
