The ongoing novel coronavirus disease 2019 (COVID-19) pandemic has been caused by the rapid spread of severe acute respiratory syndrome-2 (SARS-CoV-2). Human beings are known to be the largest reservoir of this virus. The original strain of the virus has undergone mutations, and as a result, several SARS-CoV-2 variants have emerged. Previous studies have indicated the risks of this virus of acquiring additional reservoirs, e.g., minks. This could elevate the risk of further mutations, which could result in the emergence of more infectious variants that are unresponsive to existing COVID-19 vaccines.
SARS-CoV-2 infection in mice
Scientists had previously reported that both the original strain of SARS-CoV-2 as well as its newly emerged variants did not infect mice. This is because mice's angiotensin-converting enzyme-2 (ACE2) receptors cannot bind with the viral spike protein. Thereby, the virus cannot enter mice cells.
This has caused some hindrance in COVID-19 research because the mouse (Mus musculus) is a commonly used animal model to study infection. Therefore, the virus had to be modified via various techniques, e.g., sequential passaging in mouse lung tissues and modifying the receptor-binding domain (RBD).
Several other strategies are being used to infect mice with the SARS-CoV-2 virus, including sensitizing the respiratory tract via transduction with adenovirus or adeno-associated virus expressing hACE2.
Surprisingly, many recently published studies have shown some of the SARS-CoV-2 variants of concern (VoC) originating from South Africa, the UK, and Brazil can infect mice. This has led the scientific community to worry because additional reservoirs of the virus could increase the risk of producing variants. Furthermore, as the viral reservoirs, such as mice, could be in close contact with human beings, and as they cannot be vaccinated, the chances of stopping further emergence of variants could be vastly reduced.
A new study
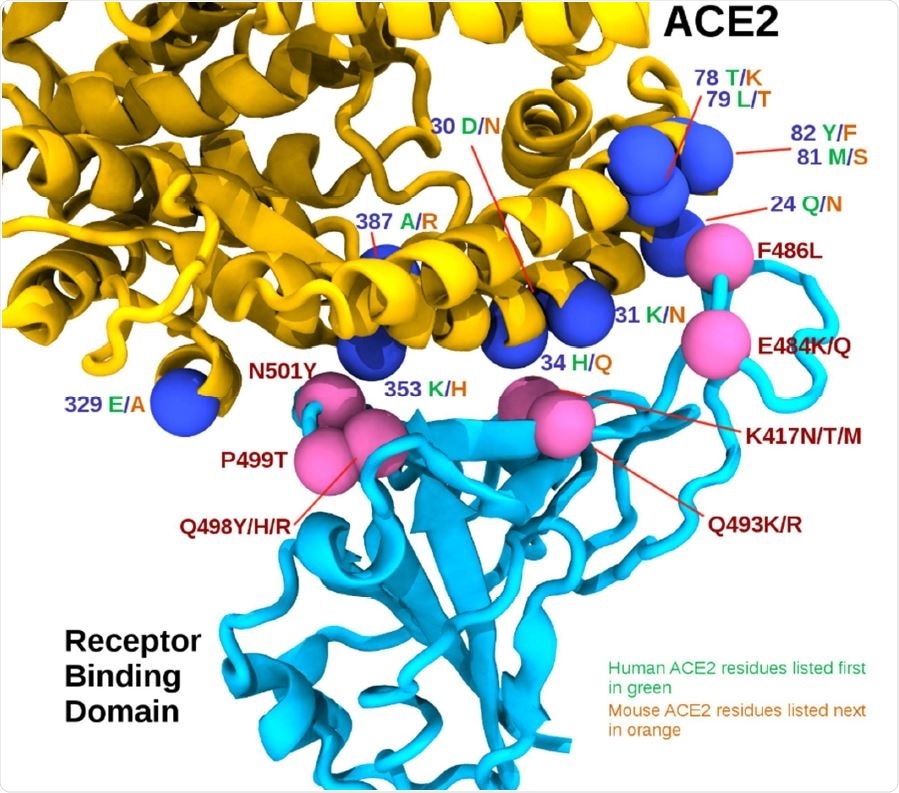
A new CSIRO study published on the bioRxiv* preprint server has conducted in silico studies along with in vitro, in vivo, and in situ observations and indicated the mouse adaptation of SARS-CoV-2. In silico studies revealed that the interface residues of the RBD/ACE2 complex in human and mouse models contain 30 ACE2 residues in close contact, of which 19 are conserved between mACE2 and hACE2.
Based on these positions, the researchers predicted that RBD mutations associated with the mouse adaptations could be grouped loosely into three regions. These are as follows:
- Region 1 (RBD positions 498, 499, and 501) located around the highly conserved ACE2 residue tyrosine 41 (Y41).
- Region 2 (RBD positions 417, 493) located around ACE2 residue 34.
- Region 3 (RBD positions 484 and 486) near the cluster of ACE2 residues 78 to 82.
The authors of the current research indicated that the mouse adaptation of the SARS-CoV-2 virus requires an electrophilic aromatic substitution, in one of the two positions of the spike protein's receptor binding domain, i.e., in position 501 or position 498. In region 1, the substitution of proline 499 to threonine (P499T) was observed in two infectious clones. Scientists further observed the binding affinity increased more owing to mutations in 417, 484, and 493 positions, e.g., K417N, E484K, Q493K, and Q493R. However, the binding affinity was reported to be less where mutations occurred in positions 486 and 499, e.g., F486L and P499T. Thereby, this research has shown that SARS-CoV-2 Alpha, Beta, and Gamma variants of concern can effectively infect mice, whereas the Delta and 'Delta Plus' cannot do so. In addition, scientists have stated that some mutations can lead to other mutations. For instance, N501Y, the primary mutation commonly present in the Alpha, Beta, and Gamma variants of concern, promote F501 mutation with a further nucleotide change. Also, E484K mutation can lead to R484 or T484 with an additional nucleotide change.
Conclusion
It is imperative to understand if the virus can adapt to a new host. A thorough interpretation of data obtained from different sources such as in silico, in vivo, in vitro, and in situ studies is essential.
The current research has revealed that the SARS-CoV-2 virus can adapt to mice, and this conclusion was drawn based on the best available evidence available at this point. The research methodology, which included the use of different approaches like bioinformatics and artificial intelligence technologies, could effectively aid future research associated with identifying other animal reservoirs and predict the probable mutation points. This approach could also help researchers predict and mitigate virus-host adaptations for the current and future pandemics. In addition, the current research has identified several countries where local field surveillance of mice is encouraged because these countries are prone to come in contact with mice bearing viruses with adaptive mutations.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Kuiper, J.K. et al (2021).But Mouse, you are not alone: On some severe acute respiratory syndrome coronavirus 2 variants infecting mice. bioRxiv 2021.08.04.455042; doi: https://doi.org/10.1101/2021.08.04.455042, https://www.biorxiv.org/content/10.1101/2021.08.04.455042v1
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: ACE2, Adenovirus, Angiotensin, Animal Model, Artificial Intelligence, binding affinity, Bioinformatics, Coronavirus, Coronavirus Disease COVID-19, Enzyme, in vitro, in vivo, Mus musculus, Mutation, Nucleotide, Pandemic, Proline, Protein, Receptor, Research, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Threonine, Tyrosine, Virus

Written by
Dr. Priyom Bose
Priyom holds a Ph.D. in Plant Biology and Biotechnology from the University of Madras, India. She is an active researcher and an experienced science writer. Priyom has also co-authored several original research articles that have been published in reputed peer-reviewed journals. She is also an avid reader and an amateur photographer.
Source: Read Full Article
